Ribosomes are the main engines of creation of the proteins on which the body depends. Now, an artificial analog of the biological ribosome has been designed and synthesized by Professor David Leigh FRS and his team in the School of Chemistry at the University of Manchester.
Organic chemists have long had the goal of making chemical-based assembler molecules that can mimic the complex biological machinery of life. In particular, ribosomes are large and complex molecular machines that make proteins (amino acid polymers) from amino acids within all living cells. Ribosomes accomplish their biological task by linking together amino acids in the order specified by messenger RNA (mRNA).
Ribosomes consist of two major subunits—the small and large ribosomal subunits. In essence, the small ribosomal subunit reads the genetic instructions on the mRNA, and directs the ordering of amino acids that will form the protein assembled by the large ribosomal unit. The power of the ribosome is that, given the raw materials, it can make any protein that can be reduced to a genetic pattern recorded on mRNA.
Leigh's group has concentrated on molecular machines that generate mechanical movement that imitates the function of macroscopic machines. A primary goal of such studies is a molecular assembler, a proposed device able to guide chemical reactions by positioning reactive molecules with atomic precision. Such a machine is potentially capable of synthesizing chemical structures that cannot be made by conventional chemical means, as the energy barriers that prevent some molecules from forming can be largely overcome through mechanical manipulation.
A number of artificial molecular machines have been fabricated in recent years. For example, small molecular machines have been used as molecular motors, to store information. Others have been used to aid the synthesis of organic chemicals to switch the action of a catalyst on and off, and to change the "handedness" of a reaction product. Large synthetic DNA molecules have been used to guide the formation of bonds between building blocks that would not normally react, and to assemble gold nanoparticles in particular sequences. However, until now there has been no analog constructed of even a crude molecular assembler.
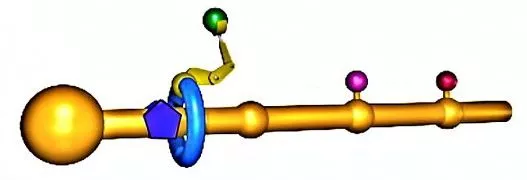
The figure above can be used to give a general notion of how the molecular assembler works (a more detailed description appears in the image gallery). Briefly, you make a polymer backbone (the yellow rod) that has attachment sites (yellow bumps) for three amino acids, and also has a large chemical group at the other end.
A ring molecule (blue ring) is positioned around the polymer backbone. The hole in the ring is large enough that the molecule can move smoothly up and down and rotate around the polymer backbone, but the ring cannot pass the large chemical group or any of the attachment sites while they are occupied by an amino acid.
A short polymer chain serves as an assembly arm (yellow "arm") that can react with an amino acid, detach it from the attachment site, and put it on the end of a protein chain. The protein chain already has three amino acids to give the arm enough flexibility to carry out this reaction in a reasonable amount of time.
As shown in the video below, in operation the ring moves up and down the polymer backbone between the large molecular group at the right and the first occupied attachment site. This motion is powered by Brownian motion, as is the assembly arm, which takes on all possible configurations on a statistical basis. Eventually, the ring is near enough to the attachment site at the same time as the assembly arm is in the right configuration to pluck the first amino acid. It does so, and the amino acid is moved by the assembly arm to the end of the growing protein chain.
The same mechanics power additional stages of polymer growth. However, the region over which the ring moves is now larger, as it its mass, and both of these factors slow down the process of adding the second amino acid to the polymer chain. Leigh finds that it takes an average of 12 hours to add each of the amino acids (there is as yet no data on how long each assembly step takes). To put this into perspective, a ribosome can add 15 to 20 amino acids to a polymer chain every second.
Prof Leigh's molecular assembler is clearly no competition for a biological ribosome. Not only is the speed roughly a million times slower, but Leigh's assembler has a hard-wired program, in contrast to the ribosome, which is programmed by the messenger RNA encoding the desired protein sequence. Despite its limitations, however, Leigh's is not only a large step in the right direction, but is also a tour-de-force of synthetic organic chemistry.
Source: University of Manchester














