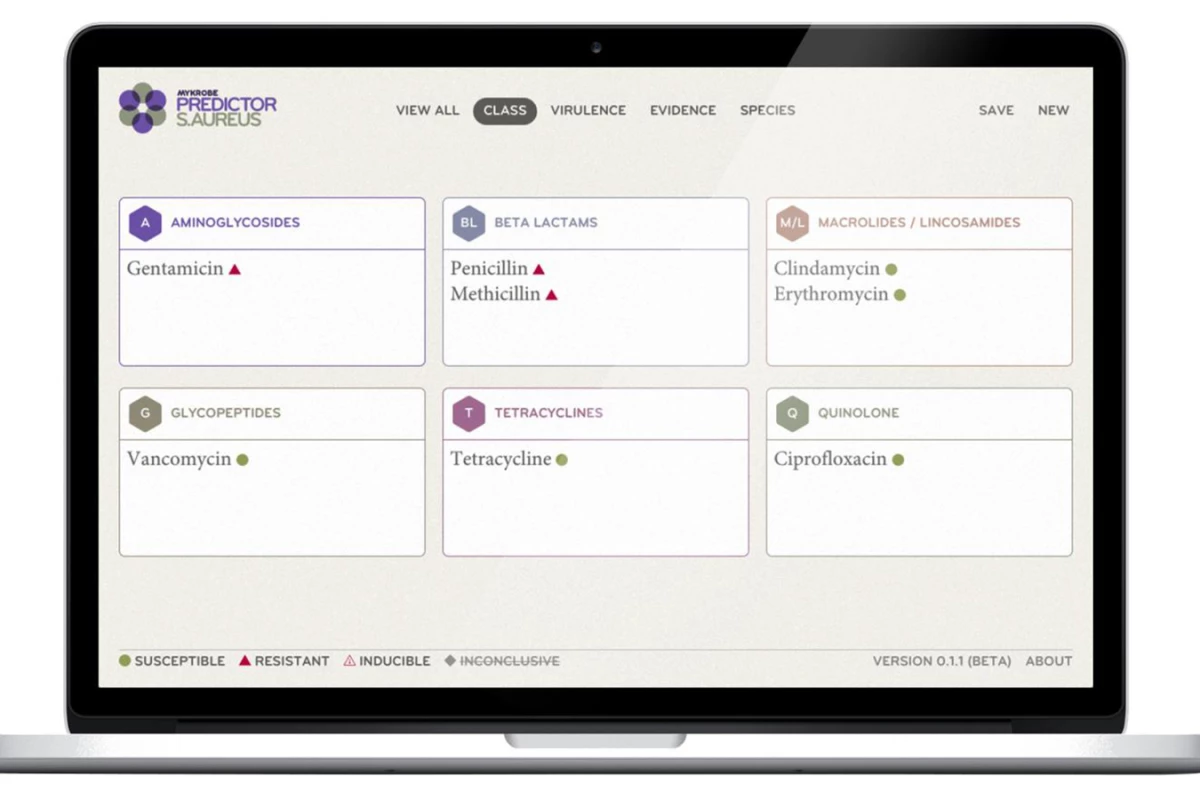
A team of University of Oxfordresearchers has developed easy-to-use software that's able to quicklypredict which antibiotics will work for a patient by analyzing DNAfrom their infection. The program is currently being trialed in threeUK hospitals.
Just like every other species on theplanet, bacteria are constantly evolving. As time goes on, some ofthe changes that occur in their DNA makes them resistant to drugsthat we use to treat them. Treatment-resistant bacteria are more likelyto survive and pass on their traits to other bacteria, creating an escalating problem, and one that could one day pose a huge risk toglobal health.
The best way to tackle the problem isto treat patients with the right antibiotic as quickly as possible,but that's not always easy to achieve. Doctors first have to identifythe bacteria that's infecting the patient, then find out whether it'sresistant to certain drugs. This is usually done by applyingdifferent antibiotics to the bacteria in a petri dish and watching tosee which drug kills it off – a process that can take days, weeksor even months in certain cases.
So, how can we speed up the process?Well, scientists think that it's possible to identify bacterias' drugresistance by searching their DNA for particular mutations that areknown to cause drug immunity. There are problems here too, however, withtypical methods requiring a lot of computing power and the presenceof an expert.
The University of Oxford researchersdecided to tackle the issue head-on, developing a computer programknown as Mykrobe Predictor, that aims to make the entire process muchmore straightforward. It allows for automated genome processing,checking bacteria DNA sequences against previously-analyzed strains,to quickly search for the mutations that cause resistance. Once theanalysis is complete, the information is presented in aneasy-to-understand manner that negates the need to have an expert onhand.
The researchers tested out thesoftware, using it to analyze more than 4,500 retrospective patientsamples, searching for antibiotic resistance in two dangerousbacterial infections – tuberculosis (TB) and Staphylococcus aureus,a form of which can cause MRSA.
The results were impressive, with theMykrobe detecting resistance to five first-line antibiotics in morethan 99 percent of Staphylococcus aureus cases – equaling theperformance of traditional methods. When testing TB samples, thesoftware was able to detect resistance in 82.6 percent of casesbetween five and 16 weeks quicker than conventional testing.
Unlike existing methods, the newsoftware can be updated as new resistance mutations are discovered,meaning it'll keep getting better as time goes on. It's also able toidentify infections even when a there are a mixture ofdrug-susceptible and drug-resistant bacteria present in a patient'ssystem.
"Our software manages data quicklyand presents the results to doctors and nurses in ways that are easyto understand, so they can instinctively use them to make bettertreatment decisions," said paper lead author Dr. Zamin Iqbal.
The software is currently being trialed in hospitals in Leeds, Brighton and Oxford, while a paper discussing it was recently published in the journal Nature Communications.
For a look at the program, you can check out the video below.
Source: University of Oxford