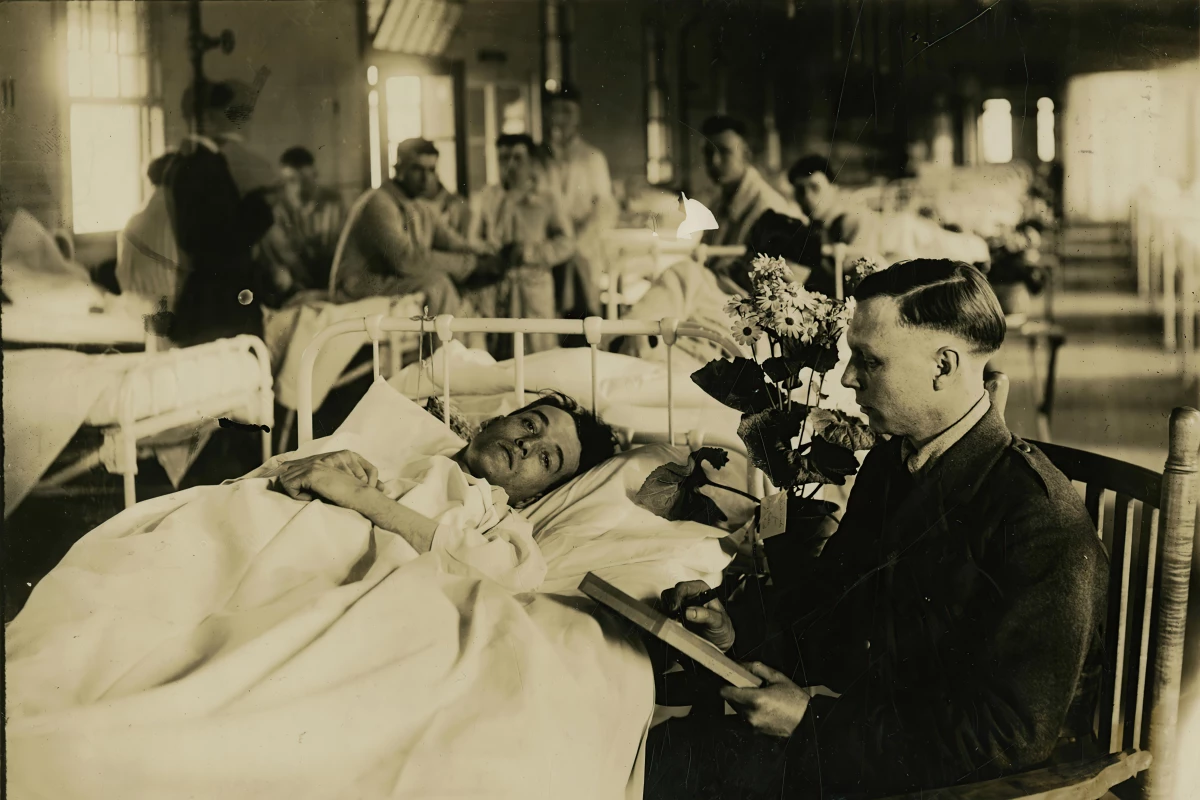
The preserved lung of an 18-year-old Swiss man has been used to create the full genome of the 1918 "Spanish flu," the first complete influenza A genome with a precise date from Europe. It offers new insights into the deadly pandemic that claimed the lives of up to 100 million people.
An international research team led by the University of Basel has applied cutting-edge technology to extract traces of the virus from the formalin-preserved organ taken from the man who died of severe pneumonia at the Cantonal Hospital (now University Hospital) in Zurich. The teen's lung had been kept in a university medical collection since his death in July 1918, during the first wave of the pandemic.
“This is the first time we’ve had access to an influenza genome from the 1918 to 1920 pandemic in Switzerland," said Verena Schünemann, a paleogeneticist and professor of archaeological science at the University of Basel. "It opens up new insights into the dynamics of how the virus adapted in Europe at the start of the pandemic."
Using a new RNA-sequencing protocol designed to extract genetic data from degraded, chemically fixed tissue, and comparing the completed genome with ones from Germany and North America, the team was able to show that this strain of the virus already had three important adaptations. These made it more deadly to humans – and they'd remain in the virus makeup until the pandemic's end.
Two of the mutations helped the virus evade a key part of the human immune system known as MxA, an antiviral protein that normally blocks influenza from replicating. This protein is especially important in defending against bird-origin influenza viruses, so these changes made it easier for the virus to spread between humans.
The third mutation altered the shape of a surface protein called hemagglutinin, which the virus uses to attach to and enter human cells. This made the virus better at recognizing and binding to human-specific cell receptors, increasing its infection efficiency.
These mutations were previously thought to emerge later in the pandemic – so their presence in Switzerland's first wave in spring 1918 suggests the virus had evolved rapidly and was widespread even before the pandemic’s second and most lethal wave in the fall.
Interestingly, the Zurich virus also showed unusual genetic diversity in its polymerase (PB2) segment, suggesting either strong natural selection or mixing between viral strains. When compared to, for example, 2009’s H1N1 virus, the 1918 bug had higher variability in key genes linked to replication and host adaptation. It also shows how quickly influenza viruses can adapt to bind to receptors in humans and evade immune system takedowns.
Not surprisingly, these rapid adaptations were also a hallmark of the coronavirus at the center of our most recent pandemic.
One of the most exciting parts of the study is the process by which the team was able to build this historic genome. Until now, this kind of wet specimen preserved in formalin had been considered unsuitable for RNA analysis. But the comprehensive genetic data that the researchers were able to extract from the lung tissue opens the door to unlocking the DNA secrets held in thousands of jars in medical and zoological collections around the world.
“Ancient RNA is only preserved over long periods under very specific conditions," said Christian Urban, the study’s first author. "That’s why we developed a new method to improve our ability to recover ancient RNA fragments from such specimens."
The researchers' ligation-based method is able to capture shorter genetic fragments, and also preserves RNA strand orientation. And by discovering the kind of adaptations seen in viruses at the center of past pandemics, researchers can gain valuable evolutionary clues that can better prepare us in tackling future outbreaks. Seeing how viruses spill over from animals to humans will also be key in developing vaccine targets.
“A better understanding of the dynamics of how viruses adapt to humans during a pandemic over a long period of time enables us to develop models for future pandemics,” Schünemann said.
The study was published in the journal BMC Biology.
Source: University of Basel