Researchers have provided experimental proof of a pathway that controls aging, finding that quieting some mobile DNA sequences in roundworms led to a longer life. Not only does the discovery provide a greater understanding of how we age, but it also opens the door to potential applications in biology and medicine.
Transposable elements (TEs), also known as transposons or ‘jumping genes’, are sequences of DNA that move, or jump, from one location in the genome to another. These moves can sometimes create mutations in the new location, causing genomic instability that influences and even promotes the process of aging and age-related diseases.
Researchers from Eötvös Loránd University (ELTE) in Hungary, had, in 2015 and 2017, published studies theorizing about how a specific process, called the Piwi-piRNA pathway, contributed to aging by helping to control TEs. Now, in their latest study, they provide experimental proof of how the pathway works.
The Piwi-piRNA pathway – whose full name is P element-induced wimpy testis in Drosophila-Piwi-interacting RNA pathway – is a specific RNA silencing mechanism that protects genomes from the adverse mutagenic activity of TEs. While the pathway acts in non-aging cells, it doesn’t act in aging somatic cells, which, due to their accumulating cellular damage, undergo a functional decline called senescence and eventually die. Somatic cells are any cells of a living organism other than reproductive cells.
In the current study, the researchers performed experiments on Caenorhabditis elegans, often used as a model system for studies on aging, age-related diseases and mechanisms of longevity because C. elegans possesses homologs (genes similar in sequence) of about two-thirds of all human disease genes.
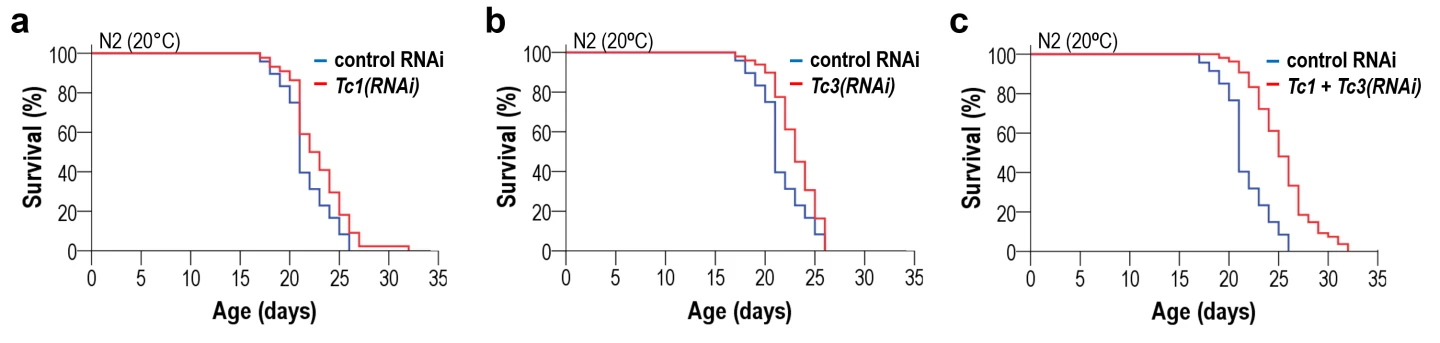
They downregulated active TE families and found that the downregulation of two specific families, Tc1 and Tc3, the most mobile TEs in the roundworm, slowed the aging process at different temperatures. At 68 °F (20 °C), lifespan was extended by around 10%. Simultaneously downregulating both caused the life-extending effects to add up to nearly the sum of lifespan expansion observed in the single treatments. Downregulation of other TE families – Tc2, Tc4, and Tc5 – produced no detectable impact on lifespan.
“In our lifespan assays, by merely downregulating TEs or somatically overexpressing the Piwi-piRNA pathway elements, we observed a statistically significant lifespan advantage,” said Ádám Sturm, the study’s lead author. “This opens the door to a myriad of potential applications in the world of medicine and biology.”
Additionally, the researchers found epigenetic changes in the DNA of these worms as they aged, specifically in the TEs. Epigenetic changes, unlike genetic changes, are reversible and don’t change the DNA sequence but can change how the body reads that sequence. DNA methylation is one of the most important epigenetic modifications and plays a key role in a number of genetic processes. Here, the researchers observed that DNA N6-adenine methylation progressively increased TE jumping as the worm aged. They determined that it may be possible to use this epigenetic modification as an accurate determinant of biological age.
“This epigenetic modification may pave the way for a method to determine age from DNA, providing an accurate body clock,” said Tibor Vellai, corresponding author of the study.
Better understanding the pathways that control aging may lead to developing ways of extending life and improving health in our later years, the researchers say.
The study was published in the journal Nature Communications.
Source: Eötvös Loránd University






